Welcome to the website of the LINK-Group!
The LINK-Group was formed in 2004 by researchers and research students from different fields and different universities to discover the topology and dynamics of networks. The group focuses on signaling and protein-protein interaction networks investigating the development of tumors and drug resistance. The LINK-Group develops novel algorithms to analyze network topology and dynamics and uses them to characterize the adaptation of complex systems to novel environments, and to identify new drug targets.
LINK Group member, Márk Kerestély MD received the Pro Scientia medal, the highest possible Hungarian award for a former university student involved in scientific research

June 2025
Our member, Márk Kerestély MD a PhD student of the LINK-Group received the Pro Scientia gold medal, an award established in 1988 for the best university student scientists of Hungary. Márk is in his 3rd year of MD-PhD studies working on dynamic network models of cell division and cancer.
LINK-Group member, Klára Schulc received an Excellent Research Student of the Semmelweis University award

February 2023
Our LINK-Group member, Klára Schulc received the highest award of research students at the Semmelweis University (Budapest, Hungary), the Excellent Research Student award. Klára is now continuing her studies as a PhD student, making progress in her research at the University of Heidelberg during this academic year.
The LINK-Group member, Erik Seitz won the 2021 Hungarian Innovation Contest and the best medical project prize of the 2021 EU Contest for Young Scientists

September 2021
Our LINK-Group member, Erik Seitz a high school student of the Pannonhalma Benedictine school won one of the four 1st prizes of the 30th Hungarian Innovation Contest. He participated in the development of a two-cell dynamic network model of epithelial-mesenchymal transition and discovered a potential drug target inhibiting this transition playing a key role in metastasis formation. Erik was working with Nina Kunsic and other members of the group. Erik also won the best medical project prize of the 2021 EU Contest for Young Scientists. Congratulations to all participating in this exciting project!
Our new paper on the prediction of activation/inhibition and sign-balance of the Caenorhabditis elegans chemical synapse neuronal connectome network has been published in PLoS Computational Biology.

June 2020
It is our great pleasure that our new method to predict the polarity (activation or inhibition) of the C. elegans connectome, incorporating presynaptic neurotransmitter and postsynaptic receptor gene expression data (3,638 connections and 20,589 synapses total) has been published in PLoS Computational Biology. The methodology can be accessed at this website: http://elegansign.linkgroup.hu Supplementary Data can be viewed here: http://linkgroup.hu/elegansdata.php. In the paper we show that approximately 20% of C. elegans connectome connections are inhibitory, which is a ratio found in many real-world networks (including partial connectomes) before. Our method opens a way to include spatial and temporal dynamics of synaptic polarity that would add a new dimension of plasticity in the excitatory:inhibitory balance.
Our new concept on the learning of signaling networks became the cover story of the 2020 April issue of Trends in Biochemical Sciences

January 2020
It is our great pleasure that our new concept demonstrating that the general rule of neuronal Hebbian learning is valid to the learning of signaling networks of single, non-neuronal cells has been published in Trends in Biochemical Sciences (accessible here and as a pdf file here) and became the 2020 April cover story of the journal. In the paper we summarize the molecular mechanisms of cellular learning, including conformational memory of intrinsically disordered proteins and prions, signaling cascades, protein translocation, RNAs (microRNA and lncRNA), and chromatin memory.
We published EntOptLayout, the only network visualization method which is able to clearly discriminate network modules

August 2019
Our network visualization method EntOptLayout uses the novel network representation theory we published in 2015. We published the EntOptLayout Cytoscape plug-in in Bioinformatics, which is the first network visualization method able to clearly discriminate between network communities. The efficiency of the methods was proved on various real world protein-protein interaction and signaling networks, as well as on synthetic benchmark graphs.
We published Translocatome: a database of experimentally verified and predicted translocating proteins was published in Nucleic Acid Research

October 2018
Our new database, Translocatome was published in the 2019 Database issue of Nucleic Acid Research (https://doi.org/10.1093/nar/gky1044) and is accessible here. The database contains a gold standard core dataset of 213 manually curated, experimentally verified translocating human proteins + 1133 human proteins predicted to be translocated by high probability based on their GO and compartmentalized interactome properties. The database was the work of mainly these LINK-Group members: Péter Mendik, Levente Dobronyi, Ferenc Hári, Csaba Kerepesi, Leonardo Maia-Moço, Donát Buszlai, Péter Csermely and Dániel V. Veres.
Our compartmentalized protein-protein interaction database (ComPPI) was updated

July 2018
Donát Buszlai and Dániel Veres have updated the compartmentalized protein-protein interaction database (ComPPI) published originally in the 2015 database issue of Nucleic Acid Research. ComPPI 2.0 has updated localization and interactome datasets and contains more than double of the protein-protein interactions than version 1.0, having 300,000 localizations and 1.87 million interactions total.
Turbine became the best Central European startup

November 2017
Turbine became the best Central European startup + also received the best AI startup award. Turbine was founded together by 4 LINK-members, Kristóf Zsolt Szalay, the inventor of Turbine, Daniel Veres CMO and Ivan Fekete CS, and Peter Csermely as an advisor, as well as by the CEO,
A paper on a general mechanism of network-driven adaptation, learning and decision making processes

November 2017
Csermely Peter's paper on a general mechanism of network-driven adaptation, learning and decision making processes appeared on-line in BioEssays, and is accessible here, or at this web-page. The paper shows that deliberative democracy is an efficient learning strategy developed as the success of a billion-year evolution.
Turbine became the best start-up of the year

September 2017
The Turbine startup became start-up of the year on the Hungary primary of the Central European Startup contest having more than 200 applicants from Hungary. The start-up won alltogether 3 categories in the contest which is rather unprecedented. Congratulations for the growing team having several LINK-Group members! (On the picture the there is the founding team, from left to right: Kristóf Szalay who invented Turbine, Iván Fekete who established the dynamic signaling network used by Turbine, Dániel Veres Chief Medical Officer, Peter Csermely and Szabolcs Nagy, CEO.)
Turbine became the #1 health startup at Pioneers'17 having more than 4000 contestants

June 2017
The Turbine startup became the #1 health startup of the Pioneers 2017 event The event had more than 4000 contestants, whose first 500 were invited to the meeting. Turbine was selected as one of the best 50 and then won the health category. Turbine was founded by several LINK Group members, such as the originator of the idea, Kristóf Szalay CTO, Daniel Veres CMO and Iván Fekete Chief Scientist + last but not least the CEO, Szabolcs Nagy. In 2016 the startup won Bayer's accelerator contest. Currently 2 of the top 10 pharma companies are already using Turbine technology to speed up their pipeline and rationalize R&D decision making.
LINK-Group member, Eszter Arany received a grand-prize at the national round of the 2017. high school student research conference

April 2017
LINK Group member, Eszter Arany with the supervision of Péter Csermely and Aurél Prósz won a grand-prize at the national round of the 2017. high school student research conference (TUDOK). She also won the chance to present her findings at the national round of the university student research conference (OTDK). Congratulations for the hard work!
Réka and István Albert became visiting professors of the Semmelweis University spending their sabbatical at the LINK-Group

February 2017
Réka Albert and István Albert became visiting professors of the Semmelweis University and spend their sabbatical at the LINK-Group.
Awarded lectures of LINK-Group members at recent scientific conferences

February 2017
LINK Group members Eszter Arany, Levente Dobronyi, Iván Fekete, Péter Mendik, István Taisz (together with Gábor Hajdú) with the supervision of Péter Csermely, Eszter Gecse, Aurél Prósz, Csaba Sőti and Dániel Veres won several awards at recent scientific student conferences of the Semmelweis University and the Hungarian Research Student Organization. Congratulations! Keep going!
The Turbine start-up was selected from 405 applicants as one of the 4 winners of Bayer's Grants4Apps accelarator competition
September 2016
The Turbine startup company, founded by several LINK-Group members was selected as one of the four winners of Bayer's 2016 Grants4Apps competition. As part of the benefits, Turbine's founders spend 3 months in Berlin working with Bayer's experts to intitiate a joint project.
LINK-Group member, Aron Ricardo Perez-Lopez, received a silver medal at the 48th International Chemistry Olympiad (IChO)
August 2016
LINK-Group member, Aron Ricardo Perez-Lopez received a silver medal at the 48th International Chemistry Olympyad.
Congratulations, good luck at the MIT from August and keep running!
LINK-Group member, Bence Botlik, received silver and bronze medals at the European Union Science and Mendeleev Olympiads
May 2016
LINK-Group member, Bence Botlik received a silver medal at the 14th European Union Science Olympiad (EUSO)
and a bronze medal at the 50th International Mendeleev Chemistry Olympics.
Congratulations, and keep running!
Our ComPPI NAR Database paper became a highly cited paper in Web-of-Science
November 2015
Our publication on ComPPI, the compartmentalized protein-protein interaction database, published in Nucleic Acids Research Database Issue 2015 became a highly cited (top 1%) paper ranked by Web-of-Science.
A general adaptation mechanism of complex systems was published as a pair of pre-prints
November 2015
Peter Csermely summarized several years of reading and thinking in a pair of preprints describing plasticity-rigidity cycles (https://arxiv.org/abs/1511.01239) and fast-and-slow network thinking (https://arxiv.org/abs/1511.01238) as general adaptation mechanisms of complex systems from macromolecules to cellular, neuronal and social networks. The latter paper is accompanided by several video illustrations, which can be seen at this web-page: https://linkgroup.hu/networkdecisions.php.
LINK-Group members established the Turbine start-up to design effective combination therapies with artificial intelligence
October 2015
Several LINK-Group members established a biotech start-up firm, which is based on Kristóf Szalay's formerly developed Turbine network dynamics tool. Turbine’s built-in artificial intelligence methods predict how e.g. any type of cancer responds to treatment, screening millions of possible drug combinations on a cancer-specific signaling network. Please find more description at this web-site: https://turbine.hu.
EntOpt Cytoscape plug-in of a novel, entropy optimization-based network visualization method published in Scientific Reports
September 2015
The EntOpt network visualization Cytoscape 3.1 plug-in (https://apps.cytoscape.org/apps/entoptlayout) is based on the concept of "A unified data representation theory for network visualization, ordering and coarse-graining" published now by István Kovács, Réka Mizsei and Peter Csermely in Scientific Reports (https://www.nature.com/articles/srep13786). The plug-in is in beta-phase, so we look for questions and advice to improve it. We are planning to extend the plug-in with hierarchical optimization, coarse-graining and ordering functions.
17 years-old LINK-Group member, Aron Perez Lopez published a paper on the better human interactome perturbation spreading effects of targets of drugs having side effects
April 2015
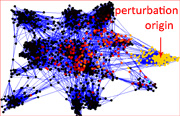
Aron Perez Lopez, a 17 years-old member of the LINK Group (https://linkgroup.hu/AronPerez.php) published a paper accepted in Scientific Reports. The paper shows that in general, drug targets are better spreaders of perturbations than non-target proteins, and in particular, targets of drugs with side effects were also better spreaders of perturbations than targets of drugs having no reported side effects in human protein-protein interaction networks. Colorectal cancer-related proteins were good spreaders and had a high centrality, while type 2 diabetes-related proteins showed an average spreading efficiency and had an average centrality in the human interactome. Moreover, the interactome-distance between drug targets and disease-related proteins was higher in diabetes than in colorectal cancer. These results may help a better understanding of the network position and dynamics of drug targets and disease-related proteins, and may contribute to develop additional, network-based tests to increase the potential safety of drug candidates.
We have updated our ModuLand plug-in for Cytoscape 3.0
February 2015
Daniel Ábrám completed and uploaded the Cytoscape 3.0-compatible updated version of our modularization program, ModuLand:
https://apps.cytoscape.org/apps/moduland20.
The ModuLand method family (PLoS ONE, 7, e12528) is determining and visualizing overlapping network modules (clusters/communities).
The plug-in builds the hierarchy of network modules, where meta-nodes of the higher layer are the modules of the lower layer.
The plug-in also calculates a number of topological measures such as community centrality, bridgeness and overlap, characterizing the centre of modules, as well as intermodular nodes (bridges) between two or more modules. More info here: www.modules.linkgroup.hu
EntOpt Cytoscape plug-in of a novel, entropy optimization-based network visualization method
January 2015
Máté-Szalay Bekő completed and uploaded the EntOpt network visualization Cytoscape 3.1 plug-in (https://apps.cytoscape.org/apps/entoptlayout), which is based on the concept of "A unified data representation theory for network visualization, ordering and coarse-graining" published earlier by István Kovács, Réka Mizsei and Peter Csermely as a pre-print (https://arxiv.org/abs/1409.8420). The plug-in is in beta-phase, so Máté is looking for all questions and advice to improve it. We are planning to extend the plug-in with hierarchical optimization, coarse-graining and ordering functions..
Kristóf Zsolt Szalay and his Turbine toolkit won the Semmelweis Innovation Award 2015
January 2015
We are happy to announce that the winner of the annual Semmelweis Innovation Award in the PhD work category is Kristóf Zsolt Szalay and his Turbine network dynamics toolkit!
Kristóf is going to hold a lecture with the title "Dinamikus hálózatelemzés és ami mögötte van" at the Semmelweis Innovation Day.
ComPPI has been accepted for publication in the NAR Database Issue 2015
October 2014
Our paper "ComPPI: a cellular compartment-specific database for protein-protein interaction network analysis" describing the ComPPI database has been accepted for publication in the Nucleic Acids Research Database Issue 2015.
Turbine wins one of the best poster awards at Interdisciplinary Signaling Workshop 2014
July 2014
The poster "Turbine: Novel dynamic analyses of signaling (and other) networks" by Kristóf Zsolt Szalay and Peter Csermely has won the Best poster award in the Network tools and resources section of the Interdisciplinary Signaling Workshop 2014.
Best interdisciplinary signaling team award
July 2014
The team working on the topic "Can we create large-scale signaling networks?" originating from the LINK-Group at the Interdisciplinary Signaling Workshop 2014 has won the Best interdisciplinary signaling team award!
Members of the group were:
- Andrei Zinovyev (team leader, Institut Curie)
- Katja Rybakova (presenter, UC Dublin)
- Kristóf Zsolt Szalay (topic owner, LINK-Group)
- Rune Linding (DTU)
- Ana Mitrovic (University of Ljubljana)
- Dénes Türei (NetBiol)
- András Zeke (ELTE TTK)
- Leilei Zhong (MIRA Institute)
More than 100,000 visitors of the LINK-Group web-site
June 2014
We would like to thank to our more than 100,000 visitors at this web-site.
We continue to add new algorithms, resources, papers, lectures and other information to the site. Please share your comments with us here: https://linkgroup.hu/email.php
Description of Turbine 1.4, an extensive tool for the analysis of network perturbations and dynamics was published in PLoS ONE
September 2013
Turbine 1.4 is an open-source network analysis package for simulating network dynamics. It is particularly suited for experiments involving perturbations (i.e.: observing the effects of outside stimulations on a target network). Turbine is written in C++, and is currently available for Windows and Linux systems. Turbine is able to accommodate any model or real world networks, single, multiple, or continuous perturbations with custom-made delay or dissipation dynamics. You may see a more detailed characterization of the program and download all its components, source code, and detailed instructions here.The PLoS ONE paper describing the results obtained by Turbine can be accessed here: Download the document! 
Description of NetworGame 2.0, a program package to simulate spatial games on model and real world networks with individual starting strategies of any nodes was published in PLoS ONE
July 2013
The NetworGame program package is a cross-platform, multi-threaded, generic tool to simulate repeated spatial games. NetworGame accommodates any model or real-world networks and is able to set individual starting strategies to any nodes. The NetworGame program package is a console application augmented with a Graphical User Interface, and a data mining tool, which helps extracting and storing data in comma-separated value (csv) format. You may see a more detailed characterization of the program and download its Linux- or Windows-compatible packages, and detailed instructions here. The PLoS ONE paperdescribing the results obtained by NetworGame can be accessed here:
https://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0067159
Our comprehensive review on the use of networks in drug design was published in Pharmacology & Therapeutics and was recommended 2 times as an F1000 Prime paper
May 2013
A comprehensive review on the topology and dynamics of networks was published in Pharmacology & Therapeutics and was recommended 2 times at the maximum, 3-star level as an F1000 Prime paper. ![]()
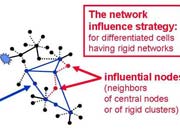
In the review we cited 1270 references, and gave extensive examples on the use of network approach in drug design. Several novel concepts were formulated, including the concept of the "central hit strategy", which selectively targets central node/edges of the flexible networks of infectious agents or cancer cells to kill them. On the contrary, the "network influence strategy" works against other diseases, where an efficient reconfiguration of rigid networks needs to be achieved. The review can be downloaded from here.
Peter Csermely was elected as a member of the Hungarian Academy of Sciences
May 2013

The LINK-member, Peter Csermely was elected as a member of the Hungarian Academy of Sciences. The Academy was established in 1825, covers all areas of science and the number of its members younger than 70 years can not be higher than 200.
Our extended signal transduction network and database, SignaLink 2.0 was highlighted by Science Signaling
March 2013

SignaLink 2.0, the recently published extension of our former signaling network and database was highlighted in the March issue of Science Signaling. SignaLink 2.0 is the second network resource from our lab, after ModuLand featured by Science Signaling and included to their selected list of bioinformatics resources.
Peter Csermely was elected as a member of Academia Europaea and as president of the European Council for High Ability
September 2012
LINK-Group member, Peter Csermely was elected at the Bergen-meeting of the Academia Europaea as a member of the academy.
He was also elected as the president of the European Council for High Ability between 2012 and 2016. The Council promotes network building of talent support acitvities in all European countries. This work is supported by the Budapest Centre of Talent Support established in the spring of 2012.
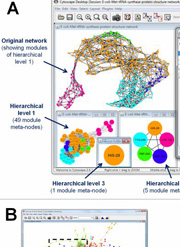
A Cytoscape plug-in of the ModuLand modularization program was published in Bioinformatics
August 2012
An updated version of the user-friendly, platform independent implementation of the LinkLand method of the ModuLand method family published earlier in PLoS ONE became available as a plug-in for the widely used Cytoscape program. The paper introducing the plug-in was published in Bioinformatics and can be accessed here. The introducing paper demonstrates the usefulness of the method
- to identify an extensively overlapping modular structure;
- to define a modular core and hierarchy allowing an easier functional annotation;
- to identify key nodes of high community centrality, modular overlap or bridgeness in protein structure, protein-protein interaction and metabolic networks.
The program file and its User Guide can be downloaded from here.
A joint paper with Ruth Nussinov on the new drug-design paradigm of allo-network drugs was published as the cover story of the December issue of Trends in Pharmacological Sciences
November 2011
The paper proposed that the concept of allosteric drugs can be broadened to allo-network drugs, whose effects can propagate either within a protein, or across several proteins, to enhance or inhibit specific interactions along a pathway. Allo-network drugs can achieve specific, limited changes at the systems level, and in this way can achieve fewer side effects and lower toxicity. The paper summarized possible methods to identify allo-network drug targets and sites. The concept outlined a new paradigm in systems-based drug design. The paper, which was published as the cover story of the December issue of Trends in Pharmacological Sciences, can be downloaded from here.
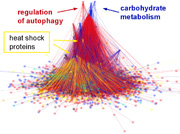
Ágoston Mihalik's paper showing a partial desintegration of the yeast interactome as a general model of systems level adatptation processes has been published in PLoS Computational Biology
July 2011
The paper described that heat shock induces a marked decrease in the overlap of the communities (modules) of the yeast interactome. Our results indicated that heat shock induces a partial disintegration of the protein-protein interaction network. The residual inter-modular bridges, maintaining the integrity of the interactome after stress, were key proteins of stress-survival. The changes observed may be rather general occurring at the initial phase of crises in many complex systems, such as proteins in physical stretch, ecosystems in abrupt environmental changes or social networks in economic crisis. De-coupling and re-wiring of modules emerges as a general model of adaptation and learning. The paper and its Supplement, which were published in PLoS Computational Biology, can be downloaded from here.
The Hungarian version of the book Weak Links was voted to the best 500 Hungarian books
June 2011
A wide panel of experts has recently determined the “Márai-list”, which is a list of the top 500 Hungarian books (including the top 250 non-fiction books) among a competing 8,000 books recommended to all Hungarian libraries. The Hungarian version of the book Weak Links (a 2009 Springer book on networks, which is downloadable from here or at Google) has been included to this selection of the top 250 non-fiction Hungarian books.
A summary paper of our results appeared in Science Signaling: Network-based tools in the identification of novel drug-targets
May 2011
A summary paper and an accompanying slide show of our work appeared in the May 2011 issue of Science Signaling. Slides were updated from the opening presentation at the International Conference on Systems Biology of Human Disease (SBHD) in Boston, Massachusetts, 16 to 18 June 2010. The paper contains several novel ideas on network dynamics and drug design, a summary of our very recent results as well as a brief description of a few ongoing projects. The paper is available here and the accompanying slideshow is downloadable from here.
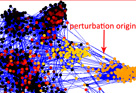
Turbine, a versatile program package for the analysis of network dynamics, including the effect of single and multiple perturbations. More...

ComPPI:
a subcellular organelle specific protein-protein interaction database with an integrated subcellular localization dataset. More...

EntOpt:
A network visualization Cytoscape 3.1. plug-in, based on the optimization of the relative entropy of the original data and their network representation More...
ModuLand:
Analysis of complex, real-world networksup to several million nodes, identification of key nodes and prediction of behavior. More...
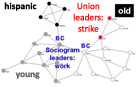
NetworGame,
a tool for the analysis of a large variety of spatial games on model and real world networks including the option for individual starting strategies for any network nodes.More...

This is the QR-code of the LINK-Group web-page. We thank you if you use it to direct the attention to this web-site at public occasions.

Questions | Address | Design and maintenance | Visitors since 7th July 2004: